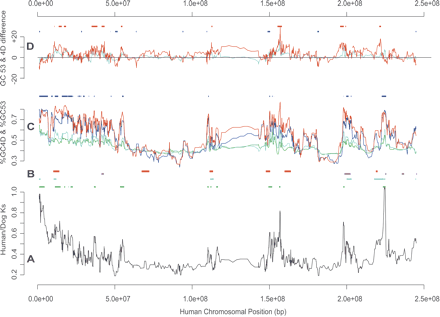
Variations in dog and human Ks, and different G+C fractions, as functions of distance (in base pairs) along human Chromosome 1. These quantities are shown as median values for 10 gene overlapping windows (see Methods). (A) The variation in Ks values (in black) of human and dog orthologs along this chromosome. Ks value hotspots are indicated in green above A. (B) The syntenic locations (see Methods) of dog telomeres (in red) and dog centromeres (in pink) on human Chromosome 1. Short synteny blocks (<4 Mb) are indicated below in light blue. (C) Variations in GC53 or GC4D fractions (as percentages). %GC53 and %GC4D values of human genes are shown in green and dark blue, respectively, whereas %GC53 and %GC4D values of their dog orthologs are shown in light blue and red, respectively. %GC4D hotspots, exceeding the 80th centile for the whole chromosome (see Methods), are marked above C in dark blue. (D) The differences in %GC53 (ΔGC53, green), and in %GC4D (ΔGC4D, dark blue), between dog and human orthologs. Above D, ΔGC4D hotspots (i.e., dog regions elevated in GC4D, with respect to human orthologs; see Methods) are indicated in red, whereas ΔGC4D cold spots (i.e., human regions elevated in GC4D, with respect to dog orthologs; see Methods) are shown in blue.











