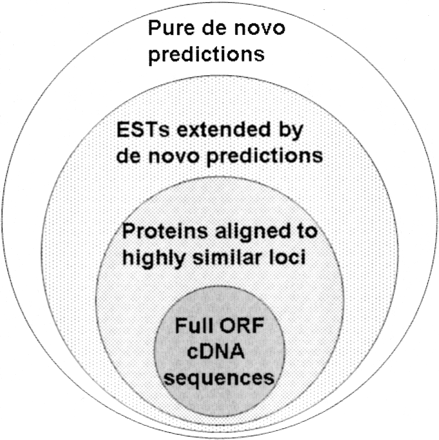
Recommended preference order for gene structure predictions. Alignments of full ORF cDNA sequences to the loci from which they were transcribed should take precedence. At loci where there is no full ORF cDNA alignment, alignments of highly similar proteins (probably >95% identity) should be used. Third best is a gene structure that is partially determined by an EST aligned to its native locus, possibly extended to a full ORF by de novo prediction. At loci where none of these are available, pure de novo predictions using dual- or multi-genome prediction algorithms should be used.











