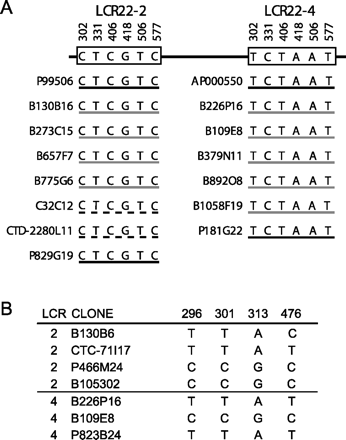
Experimental confirmation of selected PSVs and SPSs examples. (A) Paralogous sequence variants in LCR22-2 and LCR22-4 in the GGT pseudogene loci. Interval 17132218–17132600 on LCR22-2 was chosen because it contained PSV sequences identified in Figure 2. The PCR primers (F, TAGTCAGCATCAAGGTGGAG; R, CAGCACAGTAGTAGC GGATTT) amplified a 383-bp segment from the GGT locus, 4 kb downstream from the 3′-UTR. Clones were selected from different genomic libraries (genome assembly, black; RPCI PAC library, black; RPCI 11, gray; CTD library, dashed line; LL22NC03 cosmid library, C32C12, dashed line) mapping to LCR22-2 and LCR22-4 identified from the genome assembly, BAC end pairs, GenBank, and a previous report (Edelmann et al. 1999a). This report described a 4.4-kb resolution physical map of genomic clones spanning LCR22-2 and LCR22-4. This map was constructed experimentally with pure genomic clone DNA. Clones P99506 (AC008132), B130B16, B273C15, C32C12, and P829G19, for LCR22-2 and clones B226P16, B109E8, B379N11 (AC008018), and P181G22 for LCR22-4 were integrated into this map. All these clones were experimentally anchored to their respective LCR22s. Clone B657F7 is anchored to LCR22-2 because its 3′-end is in unique sequences outside the LCR. CTD-2280L11 is anchored to LCR22-2 because its 5′-end, oriented as it is, lies in the junction region of dupA and dupB (Fig. 1). The 3′-end is in dupC. This pattern is unique. It is a similar situation as described for the sequenced clone, AC007981. The 5′-end of B1058F19 is in unique, non-LCR sequences; thus, it is correctly anchored to LCR22-4. Thus, only B775G6 and B892O8 cannot be unequivocally placed into LCR22-2 or LCR22-4, but were placed in their respective LCR22s computationally (UCSC browser; http://genome.ucsc.edu/). The presumed ancestral locus in LCR8 contains TCCTAC (Supplemental Fig. 2AS). (B) Shared polymorphic sites in LCR22-2 and LCR22-4 in the GGT pseudogene loci (Fig. 2). Interval 17149097–17149647 was chosen for analysis of shared polymorphic sites. This region is duplicated in LCR22-4 and LCR22–8. The PCR primers (F, TGCCTGTTGAAAAGGCAGGA; R, CAGGCTGGCCTTTGC CAG) amplified a 551-bp segment from the GGT locus. This interval contains SPSs in genomic clones obtained from different libraries: (B) RPCI 11; (P) RPCI PAC library; CTC library. The putative ancestral locus in the GGT genomic interval contains CTAC at the same sites (Supplemental Fig. 2BS).











