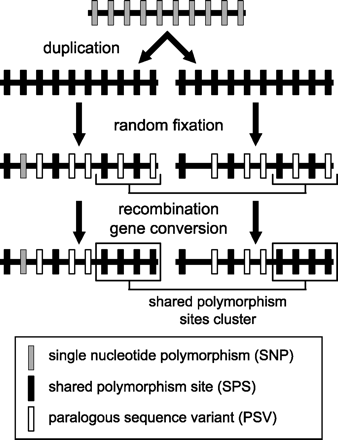
Simplified model of evolution of polymorphic sites after duplication. After a duplication, SNP sites are randomly fixed in the duplicated segments. In some cases, the process can fix the same nucleotide in both LCRs, the result being identity. In other cases, only one position is fixed and the second remains polymorphic, the result being an LCR-specific SNP (gray box). If both duplicated positions are fixed, but a different nucleotide in each case, the result is a PSV (white box). If both positions remain polymorphic, they will be detected as a shared polymorphism (black box). If the process of fixation was more or less random, we would expect to find PSVs interspersed with SPSs. This corresponds to the pattern expected from ancestral polymorphism. Recombination, on the other hand, produces separated clusters of SPSs depleted in PSVs. If the two LCRs occasionally exchange information in some individuals, all PSV positions become shared polymorphic positions (bottom), because some individuals in the population will have the original variant, and some will have a new variant transferred from the second LCR. In our model, we considered that polymorphism is transferred to both LCRs (possibly via initial gene conversion between identical segments). If only one copy remains polymorphic and the second LCR has no initial polymorphism (simple duplication event), no ancestral shared polymorphic sites are created and SPSs are solely results of other post-duplication processes (recombination). The model is simplified because we do not take into account de novo polymorphism by mutations in individual LCRs. Again, this random polymorphism would produce an interspersed pattern of PSVs and SPSs, not separate clusters.











