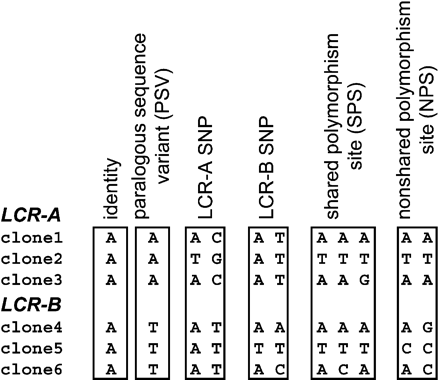
Classification of the sequence variants. Identical positions are invariant (fixed) in all clones from both LCRs. Paralogous sequence variants (PSVs) are positions invariant within each of the LCRs, but different between the LCRs. LCR-A and LCR-B SNPs correspond to positions where one LCR is polymorphic, but the second LCR is not. Shared polymorphism sites (SPSs) correspond to positions where both LCRs are polymorphic and the set of possible variants in one LCR is equal to, or contained within, the set of possible variants at the same positions in the other LCR. If both LCRs are polymorphic at a given site, but the clone variation does not overlap between the LCRs, this position is described as a nonshared polymorphism site (NPS).











