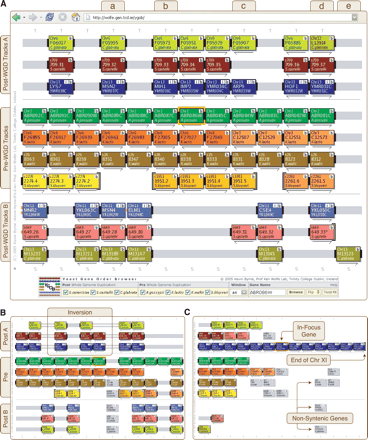
Yeast Gene Order Browser (YGOB) screenshots with a window size of six. Each box represents a gene; each color, a chromosome. The gene in focus is highlighted by an orange border. Connectors join nearby genes: a solid bar for adjacent genes, two bars for loci less than five genes apart, and one bar for loci <20 genes apart. The connectors are extended in gray over intervening space. The end of a chromosome or contig is denoted by a brace. Arrows denote relative transcriptional orientation. The “b” buttons open a window with BLASTP results against YGOB's database, “+” buttons output YGOB data in a tabulated format, “S” buttons click through to a pillar's protein sequences, and “T” buttons draw approximate phylogenetic trees on the fly. Tracks are labeled at left. (A) YGOB focused on the A. gossypii gene ABR086W. The control console at the bottom of the interface allows users to select the window size and the gene to focus on. (B) An inversion in track A of the post-WGD species, marked by orange connectors. (C) The region beside the telomere on the left arm of S. cerevisiae chromosome XI.











