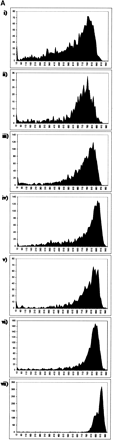
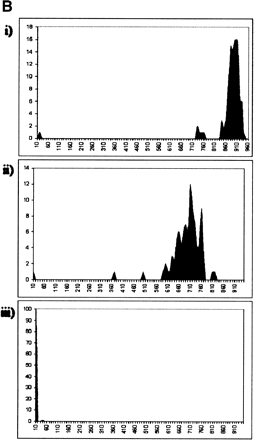
(A) Distribution of read lengths (PHRED20 base count) for each library described in Table 1, sequenced with a 400-nL reaction that contained 31.3 nL of Big Dye terminators (v3.1, Applied Biosystems). (i) CA001, CD001, CE001, CL001: Fosmid end reads; (ii) GA000: BAC end reads; (iii) CN23E: medium copy-number plasmid whole genome shotgun reads; (iv) LL005-LL0017: high copy-number plasmid 5′ EST reads; (v) TX060, TX067: high copy-number plasmid transposon-mediated shotgun reads; (vi) S1881: high copy-number plasmid SAGE library reads; (vii) 10790: high copy-number plasmid 5′ EST reads from 2304 identical clones. (B) Distribution of read lengths (PHRED20 base count) for 96 identical clones from library 10790, sequenced with a 400-nL reaction that contained (i) 15.63 nL, (ii) 7.81 nL, or (iii) 3.13 nL of Big Dye terminators.











