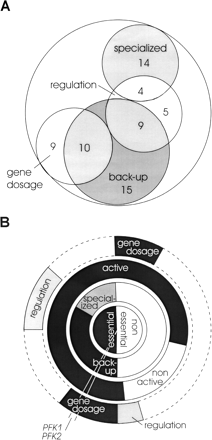
Functional categorization of the 105 duplicate gene families in S. cerevisiae metabolism. (A) Duplicate distribution of the major functions among all 105 families. The functional roles are gene dosage to boost enzyme activity, regulation by specific gene expression, back-up capability for knockout compensation, and specialized functions where knockout of one family member could not be compensated by the other(s). (B) Functional categorization of individual gene families. Each family is represented by a 3.5° angle, as indicated for PFK1/PFK2. The central circle specifies the computationally predicted essentiality of family knockouts. The first ring represents the experimentally determined single knockout phenotypes in each family. A single lethal knockout indicates a specialized function (gray) of the other(s), while viable single knockouts in a family are separated into those that clearly exhibit back-up activity (black) and those where no clear distinction can be made (white) either because they are inactive or alternative pathways could potentially compensate. The second ring specifies the experimentally determined activity of a reaction under any of the five investigated growth conditions. The third ring represents the enzyme activity modulation level. Specifically, gene dosage (black) and potentially different genetic regulation pattern (gray) can be estimated by the present data.











