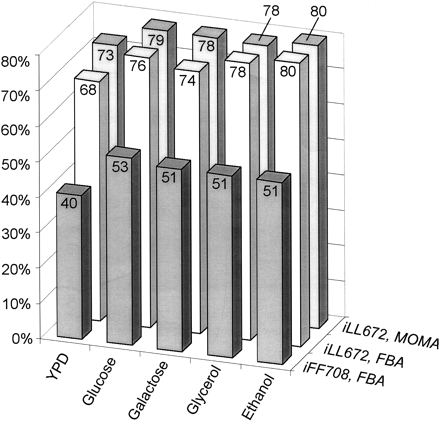
Correct prediction of S. cerevisiae mutant lethality with the in silico strain iFF708 (Förster et al. 2003a) and the in silico strain iLL672 with FBA and MoMA optimization. In the latter case, experimentally determined flux distributions were used as the reference solution (cf. Fig. 4). The sole exception was YPD medium, for which the optimal FBA solution was used. Cases in which a correct prediction is structurally not possible, that is, lethals in single-deletion mutants of duplicate gene families, were not taken into account.











