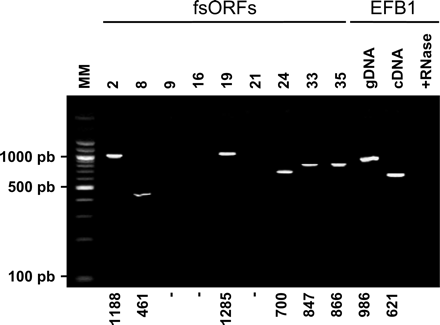
RT-PCRs. Total RNA was extracted as described in the Methods and treated with DNase I. RT-PCR was carried out in two steps. First, reverse transcription was carried out using an oligo(dT) primer, allowing only reverse transcription of poly(A) mRNAs. Then a standard PCR was performed on the mRNA after reverse transcription. The PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide. A single amplification product was seen in positive lanes; the expected size is indicated (in nucleotides) for each product at the bottom of the gel. The control sample was EFB1 mRNA, which includes an intron. Specific PCR of genomic DNA and cDNA exhibits two different products. Reverse transcription after RNase shows no DNA contamination during the process.











