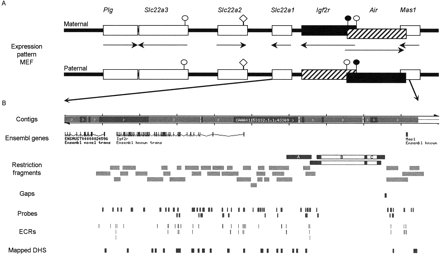
Overview of investigated region in the mouse chromosome 17 bA cluster. (A) The chromosome map spans 513 kb of mouse chromosome 17 band A and contains seven known transcripts (http://www.ensembl.org/Mus_musculus) (Lyle et al. 2000). The expression pattern in mouse embryo fibroblasts (MEFs) is shown for the maternal and paternal chromosome. Note that the 108-kb Air ncRNA overlaps the 5′ part of the Igf2r transcript by 30 kb and the 3′ part of the Mas1 transcript by 1 kb. Black boxes indicate expressed genes, striped boxes indicate imprinted silencing that affects one parental allele, white boxes indicate tissue-specific silencing that affects both alleles, arrows indicate transcriptional orientation. The Plg and Mas1 genes mark the known limits of this imprinted cluster. Note that Slc22a2 and Slc22a3 show imprinted expression in the embryonic placenta but are not expressed in MEFs. Parental-specific DNA methylation is present on the Igf2r and Air promoters. (Black circle) DNA-methylated CpG island promoter; (white circle) unmethylated CpG island promoter. Note that the Slc22a2 promoter is a weak CpG island (white diamond). (B) Modified ENSEMBL map (http://www.ensembl.org/Mus_musculus), including custom tracks (available on request) of the region including the Slc22a1, Igf2r, Air, and Mas1 genes. (Contigs row) The ENSEMBL contig view is shown, where the continuous sequence coverage stops in the 5′ region of Mas1. Note that two restriction fragments end in this region and their sizes were predicted from AJ249895. (ENSEMBL genes row) The ENSEMBL genes of this area are indicated; note that in this build Slc22a1 is annotated as “Ensembl novel trans” and the Air ncRNA is not annotated. (Restriction fragments row) Three significant differences between the public mouse sequence (C57Bl6 strain) and the sequence of the mouse strains used here (129Sv and FVB) were detected in three restriction fragments (black boxes): A—A polymorphic BclI fragment that contains a 6-kb LINE insertion only in the 129Sv mouse strain (Supplemental Fig.1); B—a 29.5-kb and C—a 6.8-kb insertion, shown in an EcoRI and an EcoRV fragment, that were not detected in any genomic DNA (see Supplemental Fig.1) and are false insertions in the ENSEMBL sequence. The positions of all other tested restriction fragments are shown as light gray boxes. (Gaps row) The gaps in the region coverage; (Probes row) DNA blot probes used for the DHS assays; (ECRs row) location of evolutionarily conserved regions (see also Figs. 2 and 3); (DHS row) the position of the identified DHS. Note that the only gap in the coverage of the region consists of a 100% interspersed repeat sequence of 1.1 kb.











