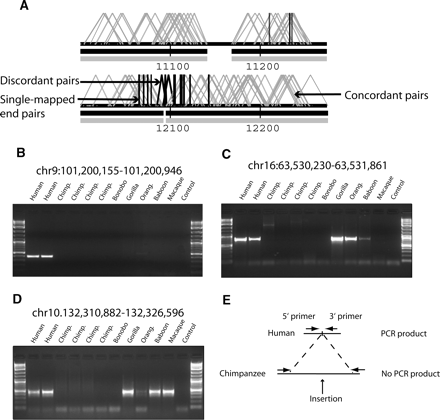
Detection and validation of “chimpanzee insertions.” (A) A chimpanzee insertion mapped to its corresponding position on human chromosome 1 (positions mapped in kb units from p arm). Two criteria were used to identify insertions: (1) two or more chimpanzee fosmids with an in silico insert size <24.9 kb (black angled lines) and (2) the presence of two or more “singletons” (vertical black lines) oriented toward the insertion. Concordant fosmid pairs are shown in gray. (B–D) PCR verification of three chimpanzee insertion events. Oligonucleotide sequences (Supplemental Table 5) were designed in regions of conserved human–chimpanzee sequence flanking each insertion breakpoint (see schematic in panel E). PCR products corresponding to the expected size were detected in human but not chimpanzees due to the increased distance between annealing oligonucleotides in the chimpanzee genome. Results from other closely related apes and Old World monkeys provide outgroup information regarding lineage-specificity of the event. Bands of unexpected size are products of non-specific binding in more distant species. (E) A schematic of the PCR primer design in chimpanzee and human.











