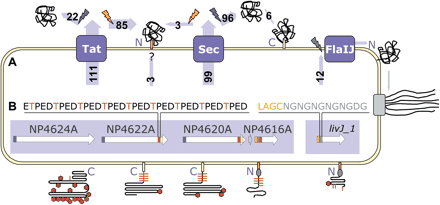
Schematic representation of protein secretion, anchoring, and glycosylation in N. pharaonis. (A) Substrates of the Tat, Sec and flagellin-specific protein translocation systems (blue boxes) are cleaved by signal peptidases (flash signs), and partly remain C- or N-terminally anchored to the cell membrane. Secreted proteins are cleaved by signal peptidase type I (blue), whereas lipobox-containing proteins are cleaved by signal peptidase type II (orange) and N-terminally attached to a lipid anchor (orange box). Lipoproteins are frequently transported via the Tat pathway (substrate numbers indicated in light-blue arrows). For three lipobox-containing proteins, the export pathway remains as yet unassigned. Furthermore, six proteins are likely to be modified by a C-terminally attached lipid anchor (yellow box). After cleavage by membrane-bound preflagellin peptidase (green), the substrates of the flagellin-specific export pathway reveal an N-terminal hydrophobic stretch possibly involved in membrane retention. (B) Signal sequence and peptide repeat modules (indicated by colored boxes) for a representative gene cluster (white arrows) are presented diagrammatically in models of the cell surface proteins. A Thr-rich tetrapeptide repeat (red box in genes, red line in proteins), likely to be O-glycosylated, occurs in several cell surface proteins adjacent to the C-terminal or N-terminal lipid anchor. An Asn-Gln dipeptide repeat (gray box, oval) follows directly after the lipobox-containing Tat-related signal sequence (orange box) of several membrane components. Other indicated features are Sec-related signal sequences (blue box in genes) and N-glycosylation sites (red hexagons in proteins).











