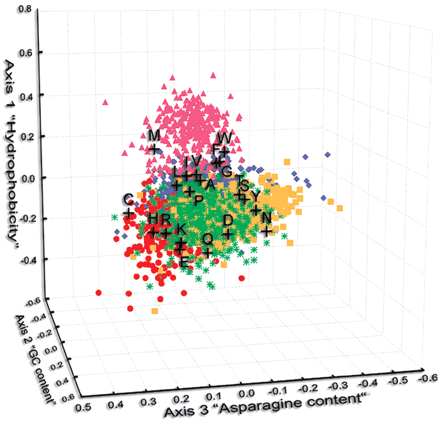
Distribution of the protein sequences on the CA space determined by the three first factors. The first one discriminates proteins by their hydrophobicity; the second one, by the GC content of genes coding for them; and the third one, by their asparagine content. Five classes of proteins are found by clustering method (see “General Features of Proteome”) and are represented: (1) IIMP by pink triangles, (2) proteins involved in the metabolism of small molecules by blue diamonds, (3) proteins associated to information transfer pathways by red circles, (4) proteins associated to the outer membrane or secreted by green stars, and (5) proteins with unknown functions, or likely to be of phage origin, by yellow squares. Amino acids are represented by black pluses.











