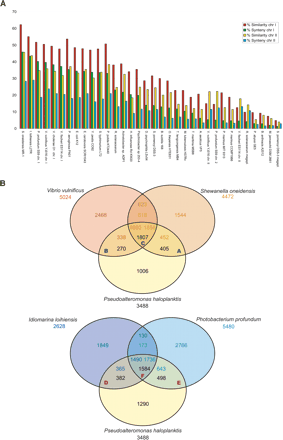
Putative orthologs and syntenies between the genome of P. haloplanktis and the genome of related bacteria. The alphabetic letters A to F refer to the Supplemental Table 4 (T4, A to F). (A) The percentage of P. haloplanktis genes homologs to a selection of 34 complete bacterial proteomes (i.e., 30% identity and a ratio of 0.8 of the length of the smaller protein to that of the larger one) is represented by red bars for chrI and by yellow bars for chrII. The percentage of P. haloplanktis genes in synteny groups with a selection of 34 complete bacterial genomes is represented by green bars for chrI and by blue bars for chrII. The closest organism is S. oneidensis. (B) Comparison of the gene content of Pseudoalteromonas haloplanktis, Shewanella oneidensis, Vibrio vulnificus, Photobacterium profundum, and Idiomarina ioihiensis. Putative orthologs are defined as genes showing a minimum of 30% identity and a ratio of 0.8 of the length of the smaller protein to that of the larger one, or as two genes included in a synteny group. The intersections between the three circles give the number of genes found in the two or three compared species. Genes outside these areas are specific to the corresponding organism. The total number of annotated genes is also given under each species name.











