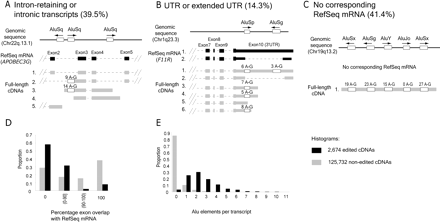
Characterization of edited transcripts. Below the genomic location is the RefSeq mRNA, in black. Exons are wide blocks, UTRs are narrower blocks, and introns are dashed lines. Alu sequences present within edited transcripts are empty blocks, and the number of A-to-G substitutions is shown above each block. Other (A) APOBEC3G locus exemplifies intron-retaining transcripts. Accession numbers for the transcripts shown are NM021822 for RefSeq mRNA, and (1) AF182420, (2) AK092614, (3) AX748234, (4) AK022802, (5) BC061914 for full-length cDNAs. (B) The F11R locus exemplifies Alu-associated editing within a UTR. Of note, the positions of edited bases for the four edited transcripts were only partially overlapping. The accession numbers are (1) NM016946, (2) NM144501 for RefSeq mRNAs, and (1) AF172398, (2) AL136649, (3) AK026665, (4) BC001533, (5) AY154005, (6) AY358896 for full-length cDNAs. (C) AK126984 exemplifies transcripts that have no corresponding known gene. This particular transcript is extensively edited in four of five Alus. (D) Deviation from known gene structure measured as percentage exon overlap with RefSeq mRNA. (E) Edited transcripts are enriched in Alu elements. The distribution of edited versus nonedited transcripts in each of the histograms was determined to be significantly different (p < 10-9).











