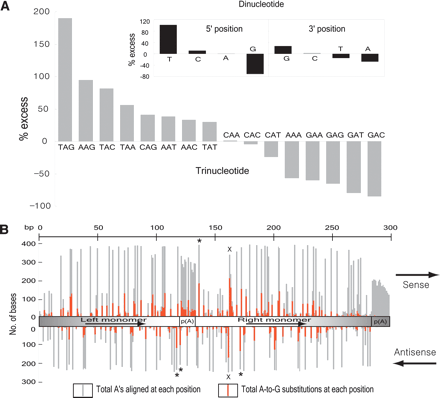
Analysis of preferred editing patterns within Alu sequences. (A) Nearest-neighbor preferences derived from 2868 Alu sequences with at least five A-to-G substitutions (n = 25,493 total edited bases) in either orientation. We determined the observed and expected frequencies of di- and tri-nucleotide patterns using the edited version of Alu sequences and their corresponding genomic sequences, respectively. We calculated percentage excess using the formula, [(observed frequency) - (expected frequency)]/(expected frequency) × 100. Previous studies (Polson and Bass 1994; Lehmann and Bass 2000) showed 5′ preferences of T ≈ A > C > G and T = A > C = G for ADAR1 and ADAR2, respectively; and 3′ preferences of nothing apparent and T = G > C = A for ADAR1 and ADAR2, respectively. (B) Identification of potential hotspots for editing within the Alus family consensus sequence, generated by separate alignments of 412 sense and 260 antisense sequences with at least 10 A-to-G substitutions using CLUSTALW (http://www.ebi.ac.uk/clustalw). We used Alus with 10 or more A-to-G substitutions to get a reasonable input size for alignment and thus minimize the effects of alignment artifacts. The number of As aligned at each position is not uniform because of sequence divergence in individual Alus. The upper panel represents the sense strand and the bottom panel the antisense strand. The basic Alu structure, as shown in the middle, consists of two nonidentical direct repeats (left and right monomer), linked by a central A-rich region. The right monomer is followed by a 3′-poly(A) that is required for Alu replication. The most frequently edited trinucleotide motif, TAG, is labeled with an *. The right monomer hotspot, present on both strands (containing a palindromic TA), is labeled with ×. Sense strands are in general more prone to editing than antisense strands, probably because of differences in availability of As (80 As and 44 As on sense and antisense Alus consensus sequence, respectively).











