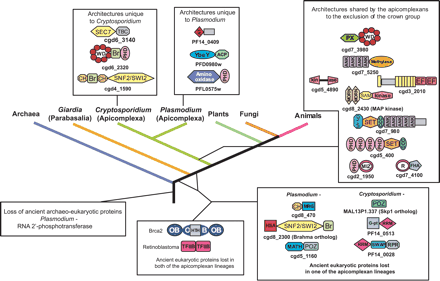
Eukaryotic tree showing select points of derivation and loss of various architectures. The proteins are designated by either Plasmodium or Cryptosporidium gene names shown below a cartoon of their architecture. Gray boxes indicate globular domains that are not detected elsewhere. Yellow boxes indicate transmembrane segments or signal peptides. The domains found in chromatin proteins are (Ch) chromo domain; (Br) bromo domain; (PHD) PHD finger; (SET) SET protein methyltransferase domain; (CCC) cysteine cluster associated with SET domains; (AT) AT hook domain; (HTH) helix-turn-helix domain; (MYB) MYB-type HTH domain; (TFIIB) TFIIB-like HTH domain; (OB) oligomer-binding domain; (SWI2/SNF2) ATPase module of chromatin-remodeling proteins; (HAS) domain found in SWI2/SNF2 ATPases. The signaling domains are MYND and MIZ-Zn-finger domains; (R) ring finger domain; (ANK) ankyrin domain; (WD) WD40 β-propeller domain; (Kinase) protein kinase domain; (SAM) sterile α-motif domain; (Sec7) ARF GTPase exchange factor domain; (TBC) GTPase-activating domain; (MORN) a β-hairpin repeat motif; (POZ) pox virus zinc finger domain; (MATH) meprin-A5-TRAF homology domain; (EF) EF-hand domain. RNA-binding domains are (G-patch) glycine-containing RNA-binding domain; (SWAP) suppressor of white apricot domain; (RRM) RNA recognition motif domain. Other domains are (YbeY) predicted metal-dependent lecithinase domain; (ACP) acyl carrier protein domain. cgd8_2430 is a predicted MAP kinase, cgd5_4390 kinase shows a lineage-specific expansion in Plasmodium, and cgd3_2010 is predicted to be a novel signaling receptor with intracellular calcium-binding EF-hand domains.











