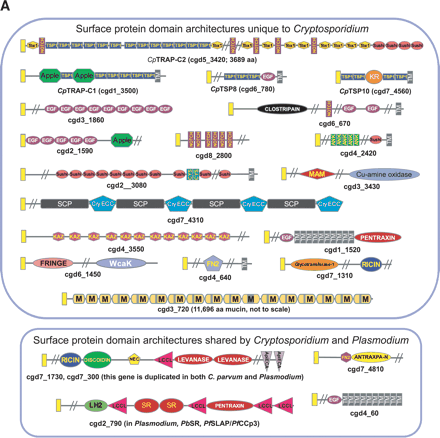
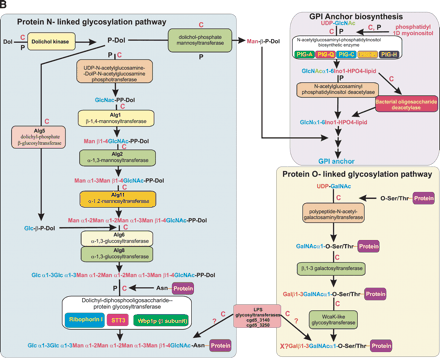
(A) Domain organizations of a representative set of surface proteins from Cryptosporidium parvum (top panel) and orthologs common to Plasmodium falciparum (bottom panel). All proteins shown here have a signal peptide sequence represented by a yellow rectangular box at the beginning of the architectures. The domains are labeled as in Supplemental Table 1. Those not shown in Supplemental Table 1 are (Ank) ankyrin repeat; (CYS) cysteine-rich repeats found in Archaeoglobus proteases; (M) mucophorin domain (with Thr/Ser stretches indicated by gray boxes; see Supplemental Fig. 1); and (TM) membrane-spanning region. (B) Schematic representation of the reconstructed glycosylation pathways in Apicomplexa. The enzymes are shown in boxes along with the protein names of the respective yeast homologs. The reconstructed oligosaccharide chain is shown using abbreviations for the various sugars. (Glc) Glucose; (Gal) galactose; (Man) mannose; (GlcNAC) N-acetylglucosamine; (GalNAC) N-acetylgalactosamine; (Dol) dolichol; and (Ino) inositol. (X?) The uncharacterized sugar added by the WcaK-like glycosyltransferases. Wherever Cryptosporidium contains an enzyme of the pathway, it is indicated with a C in red, and Plasmodium is indicated with a P in black.











