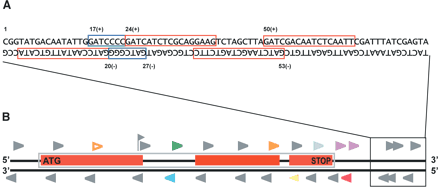
Extraction and classification of signatures from genomic sequence. (A) Example of signatures extracted from genomic DNA sequence. Red and blue boxes indicate signatures identified in the genomic sequence; a noncomplementary signature on each strand is identified from each DpnII site because of the palindromic nature of the site. Numbers above and below the sequence indicate the nucleotide position and strand information stored for each signature. Blue boxes indicate overlapping genomic signatures; the more 5′ signature can either be grouped with the 3′ signature and contain a second occurrence of GATC or be separated and consist of a signature of <17 bases or 20 bases. (B) Horizontal black lines indicate the two strands of DNA. Red boxes indicate exons of a gene on the top strand; the blue box enclosing the exons denotes the extent of the entire gene. Arrowheads indicate the positions of signatures found in the sequence. Signatures duplicated in the genome are indicated using hollow arrowheads; filled arrowheads indicate signatures unique in the genome. The format of the diagram is the same as used in the viewer on our Web page (http://mpss.udel.edu/at). Expressed signatures are indicated in color, whereas nonexpressed genomic signatures are shown in gray. The color of the triangle indicates the signature “class,” and the colors are used as follows: (orange) Class 1—in an exon, same strand as ORF; (purple) Class 2—within 500 bp after stop codon, same strand as ORF; (yellow) Class 3—antisense of ORF; (red) Class 4—in genome but not Class 1, 2, 3, 5, or 6; (green) Class 5—entirely within intron, same strand; (blue) Class 6—entirely within intron, antisense; (light green flag) Class 7—signature includes an exon/intron boundary and is spliced. Not shown are Class 0 signatures that are identified by MPSS but do not match to the genome.











