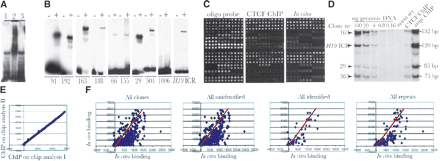
Characterization of the CTCF target-site library. (A) The bulk of the CTCF target-site library of mouse fetal liver interacts with CTCF in vitro as determined by bandshift analysis. Lane 1 depicts inserts from the library cut with NotI as probe and no protein; lane 2 shows band-shift with recombinant CTCF. The specificity of the band shift was ascertained by including a 100-fold molar excess of cold H19 ICR as competitor (lane 3). (B) Band shift assays with nine randomly picked individual clones. The first eight clones are positive, whereas clone 1006 is negative for band-shift. The – and + symbols indicate absence and presence of recombinant CTCF, respectively. (C) Microarray analysis of the CTCF target-site library. From left to right, the images show total DNA estimation using an oligo hybridization probe, pattern of in vivo (using ChIP DNA from mouse fetal liver) and in vitro (using recombinant CTCF) interactions. Each clone is represented by four adjacent spots that were printed in duplicates on each microarray. (D) The random amplification of ChIP DNA is unbiased for five randomly selected sequences. The first five lanes show the genomic DNA concentration to ascertain that the PCR amplifications were performed under semiquantitative conditions. (E) Comparison of two independent ChIP-on-chip hybridization experiments. (F) Quantitative scatterplots of in vivo/in vitro binding patterns distributed over different classes of sequences as indicated in the images. The red lines represent an estimate of in vivo/in vitro CTCF binding efficiency based on the 10 highest values.











