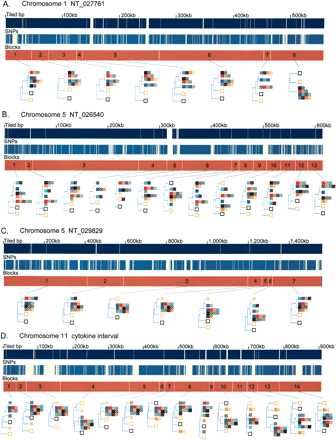
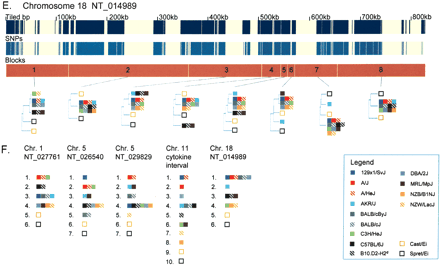
Phylogenetic relationships of the 13 inbred classical strains and the two wild-derived strains. (A–E) For each of the five sequence contigs, the phylogenetic relationships within adjacent segments are shown. (Top and middle panels) The locations of unique sequences represented on the high-density arrays and identified SNPs, respectively. (Third panel) The segmental block locations (numbered), the boundaries of which were determined by analyzing transition points between high SNP and low SNP blocks for all 78 pairwise sequence comparisons of the 13 inbred classical strains. The phylogenetic relationships of the 15 inbred mouse strains (color-coded squares), determined using the Kimura 2-parameter model, are shown for each segment. For the wild-derived inbred strains, CAST/EiJ and SPRET/EiJ, uneven amounts of missing data may affect the relative branch lengths of these outgroups in some trees as a result of unbalanced loss of CAST or SPRET specific polymorphisms. (F) For the five sequence contigs, classical inbred strains that were in the same phylogenetic group for every segment in the contig are shown together. Missing SNP data are ignored for this analysis. Related strains are depicted as solid and hatched squares of the same color (A/J and A/HeJ, BALB/cByJ and BALB/cJ, C57BL/6J and B10.D2-H2d, NZB/B1NJ and NZW/LacJ).











