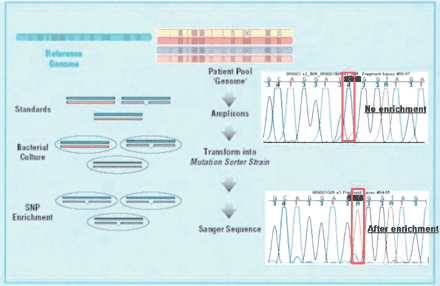
A schematic of the MRD SNP discovery process. Genomic DNA samples are pooled together, and PCR amplicons are generated by using the pooled genomic DNA as a template. If this PCR product was simply sequenced, the SNP shown would be lost in the noise of sequencing (top trace). The various amplicons (each amplified by using the genomic pool sample as a template) are pooled and hybridized to standards and transformed into the specifically designed mutation sorter strain. The fragments containing mismatches between the test sequence and the standard grow preferentially. This variant-enriched pool is then used as a template for PCR amplification and sequencing of the various amplicons. As seen when comparing the top and bottom traces, the minor allele becomes dominant after enrichment. Of note is that the frequency of this SNP is 15% in the tested population, as estimated by peak height as previously described (Kwok et al. 1994).











