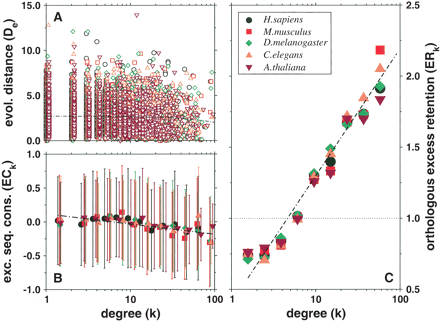
(A) Scatterplot showing for each yeast protein the evolutionary distance D to its orthologs in a higher eukaryote as a function of the proteins' number of interactions k. The thin black line corresponds to D ∼ αk with 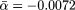 . (B) By applying logarithmic binning to the horizontal k-axis, we obtain improved correlations of the excess sequence conservation ESCk on k (〈ESCk〉 ∼ βlog k,
. (B) By applying logarithmic binning to the horizontal k-axis, we obtain improved correlations of the excess sequence conservation ESCk on k (〈ESCk〉 ∼ βlog k, 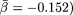 . (C) The orthologous excess retention parameter ERk shown as a function of logarithmically binned ks indicates a statistically significant monotonic trend toward the conservation of the more connected proteins (ERk ∼ γlog k,
. (C) The orthologous excess retention parameter ERk shown as a function of logarithmically binned ks indicates a statistically significant monotonic trend toward the conservation of the more connected proteins (ERk ∼ γlog k, 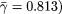 ). In each plot, the symbols correspond to the orthologs of S. cerevisiae identified in the higher organisms listed in the legends box of C.
). In each plot, the symbols correspond to the orthologs of S. cerevisiae identified in the higher organisms listed in the legends box of C.











