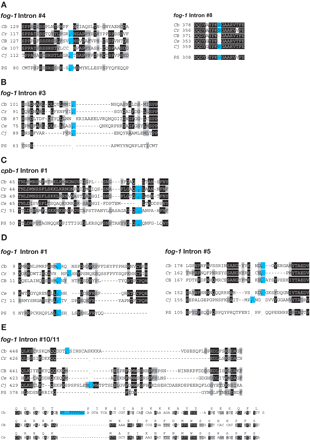
(A) Perfect deletions. (B) Deletion with 3′ insertion. (C) Deletion with associated mutation. (D) Intron sliding. (E) Intron insertions. Sequences of selected intron sites. The starting position of each sequence follows the species name. Identical amino acids are shaded black, similar ones are shaded gray, and intron sites are colored blue. A “-“is used for amino acid gaps, and a “.” for intron gaps. Phase 0 introns lie between the indicated codons, and phase 1 or 2 introns lie within the codon to the left. The bottom half of E contains nucleotide sequences, with the translation above the line. (Cb) C. briggsae; (Cr) C. remanei; (CB) C. sp. CB5161; (Ce) C. elegans; (Cj) C. japonica; (PS) C. sp. PS1010.











