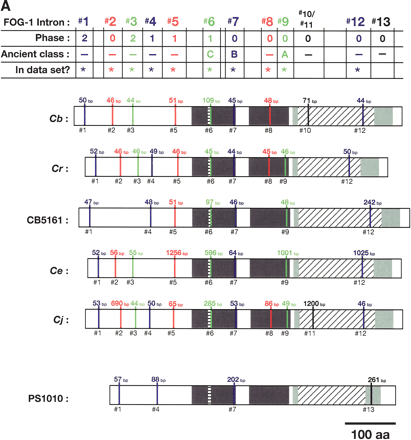
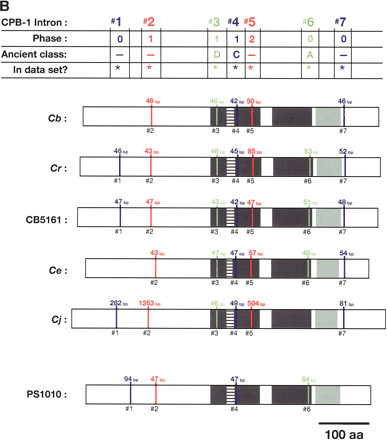
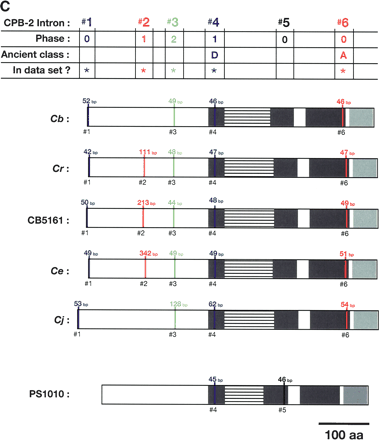
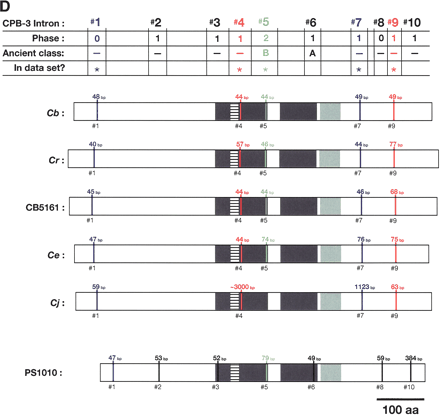
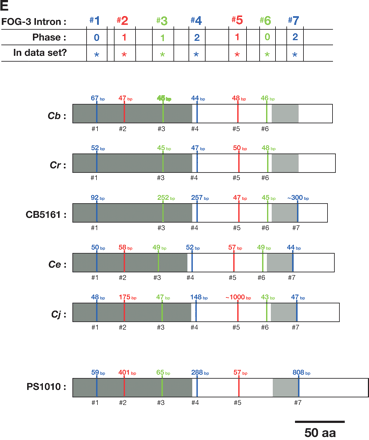
Alignment of conserved intron positions. The coding region of each gene is shown as a rectangle drawn to scale. (A–D) The RNA recognition motifs (RRMs) are dark gray, the C-H domain is light gray, insertions within the first RRM are horizontally striped, and insertions within the C-H domain are diagonally striped. (E) The BTF domain is dark gray, and the TF domain is light gray (Chen et al. 2000). Introns are shown as vertical lines within each gene, with size above, and number below. The introns used in our data set are colored blue, red, or green to make it easy to compare homologs between species. Phase 0 introns lie between codons, phase 1 introns after the first nucleotide of a codon, and phase 2 introns after the second nucleotide. Ancient introns A, B, C, and D were found in more than one CPEB gene.











