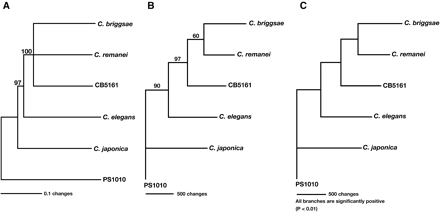
Phylogeny of the genus Caenorhabditis. (A) Neighbor-joining tree. The FOG-1, CPB-1, CPB-2, CPB-3, and FOG-3 sequences were concatenated, giving a total of 2839 characters for each species. We aligned the sequences using ClustalX, with final adjustments by hand, and used PAUP* 4.0b10 to build the tree. After 10,000 bootstrap replications, only segregations with a support value higher than 60% were accepted as significant. (B) Maximum Parsimony tree. We used PAUP* 4.0b10. Only segregations with a support value higher than 60% after 500 bootstrap replications were accepted as significant. (C) Maximum Likelihood tree. We used the Phylip software package and the JTT amino acid change model for these calculations.











