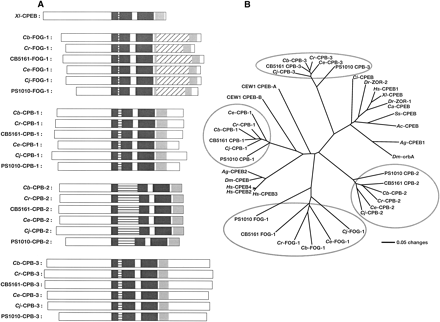
Each Caenorhabditis species has four CPEB genes. (A) CPEB domain structures. Each protein is depicted as a white box, with the amino terminus at the left. The RNA recognition motifs (RRMs) are dark gray and the C-H domain is light gray. Insertions within the first RRM are horizontally striped, and insertions within the C-H domain are diagonally striped. (Xl) Xenopus laevis; (Cb) Caenorhabditis briggsae; (Cr) C. remanei; (CB5161) C. sp. CB5161; (Ce) C. elegans; (Cj) C. japonica; (PS1010) C. sp. PS1010. (B) Neighbor-joining tree showing the relationships among CPEB genes. This tree is based on an alignment of sequences from the conserved RRM1, RRM2, and C-H domains. Each nematode gene family is circled in gray. Nematodes are listed in A, but also include Oscheius tipulae (CEW1). Other animals are as follows: (Dm) Drosophila melanogaster; (Ag) Anopheles gambiae (malaria mosquito); (Ss) Spisula solidissima (Atlantic surf clam); (Ac) Aplysia californica (California sea hare); (Ci) Ciona intestinalis (ascidian tadpole); (Dr) Danio rerio (zebra fish); (Ca) Carassius auratus (gold fish); (Hs) Homo sapiens.











