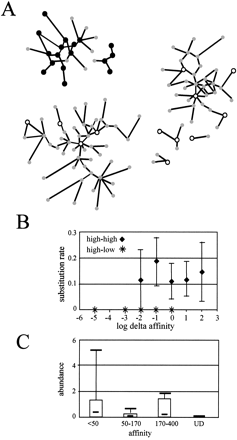
The effect of binding site affinity on Leu3 multimodality. (A) Cluster structure in a fragment of the Leu3 selection network. Black nodes, high-affinity (kd < 50 nM) motifs; white, low-affinity (kd > 170 nM); small gray, unknown affinity. Arcs connect neighbors with high substitution rate. Arcs with low substitution rates are not shown. As only motifs with measured affinities and their neighbors are presented, some real families may appear fragmented. Note that no component contains both high- and low-affinity nodes. (B) Rate of substitution as a function of affinity change within and between the families. Substitutions with similar effect on log(kd) are grouped together, and their joint rate is plotted. The rates of substitution between high-affinity sites and from high- to low-affinity sites differ significantly (P < 0.01). (C) Motif abundance box-plots for different affinity intervals. Both nonfunctional motifs (undetectable kd-s) and medium-affinity motifs (50 nM < kd < 170 nM) have very low abundances compared to motifs in the high- and low-affinity intervals, which were also identified as families in the selection network. This may indicate that medium-affinity motifs are selected against to avoid ambiguity of site modality and to increase transcriptional program robustness.











