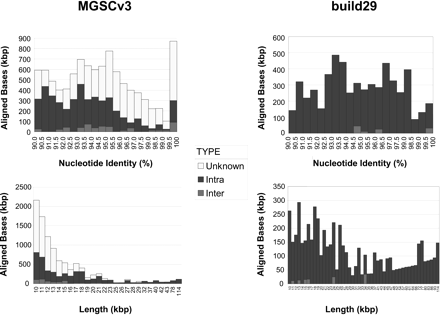
Whole-genome alignment (WGAC) statistics of the mouse draft and the build29 finished genome. Alignment statistics are binned in terms of percent identity or length (≥10 kb). We performed BLAST-based segmental duplication detection on MGSCv3 and the finished portion of build 29. The finished build 29 subset represents 439 Mb (17.7% of the draft assembly size). The abundance of aligned bases between 99.5%–100% that map to the unknown chromosome in MGSCv3 may represent highly similar duplication requiring further characterization. The build 29 pairwise were hand curated to remove uncharacterized interspersed transposable elements (Methods).











