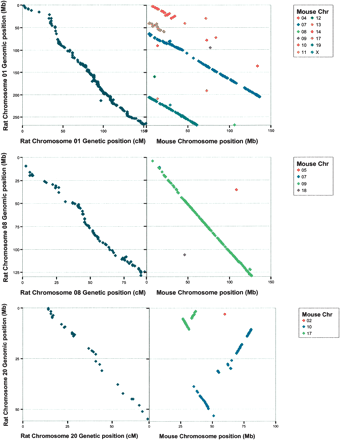
Examples of correlation between genetic and genomic positions and comparative analysis between rat (RNO) and mouse (MMU) genomes for RNO 1, 8, and 20. Each point corresponds to a rat microsatellite marker (e-PCR) or a rat EST sequence (BLAT). The genomic localization of the microsatellite markers assigned to RNO 1, 8, and 20 linkage maps derived from the GKxBN intercross was determined by the e-PCR method (left-side panels). Lengths are in cM (Kosambi) in the linkage map and Mb in the genomic sequence. Chromosomal synteny conservation was defined by BLAT query of rat EST sequences (right-side panels). The orientation of the rat chromosomes is as established by Szpirer et al. (1998). Graphs for all autosomes are available in the Supplemental material and at http://www.well.ox.ac.uk/rat_mapping_resources.











