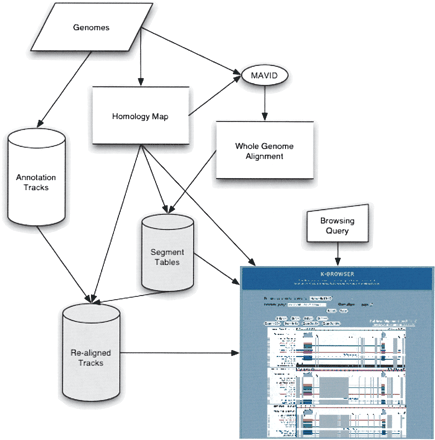
Schematic diagram of the K-BROWSER infrastructure. K-BROWSER builds two new databases (shaded) in addition to the standard annotation track database obtained from UCSC. These databases are based on a whole-genome alignment constructed using a homology map (in our case the blocks were aligned with the MAVID program). Using the segment tables and realigned tracks, a user query can be rapidly converted into a K-BROWSER figure.











