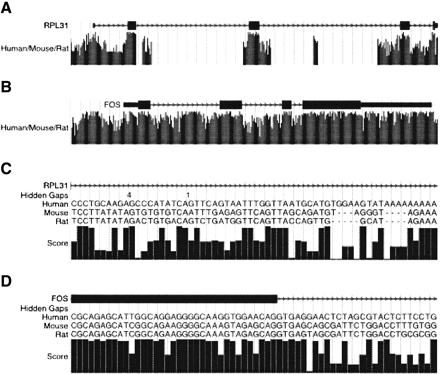
UCSC Genome Browser display of HUMOR alignments. (A) Ribosomal protein RPL31. The human/mouse/rat track shows the MULTIZ score normalized as described in the text. The high conservation of exons relative to introns is typical of many genes. (B) Transcription Factor FOS. In highly regulated genes such as this one, it is not unusual to find extensive conservation outside of protein-coding exons. (C) Closeup of a poorly conserved part of a RPL31 intron. When the display is zoomed in close enough, the base-by-base alignment is displayed as well as the score graph. Because the alignment is projected onto the reference sequence, a “Hidden Gaps” row indicates areas where in the full alignment there would be dashes in the reference sequence row. Clicking on the human/mouse/rat track takes you to a details page that displays the full alignment. (D) Closeup of an exon/intron boundary in FOS. The canonical “GT” 5′ consensus sequence is usually conserved, but then conservation falls off for the rest of the intron.











