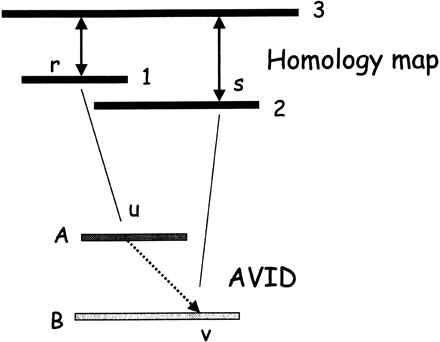
The top half of the figure shows two exon matches determined from the homology map. In particular, exon r in sequence 1 is aligned to an exon in sequence 3, and exon s in sequence 2 is aligned to another exon in sequence 3 (double arrows). At this stage, none of the sequences have been aligned - the matches are based on the pairwise protein alignments of the predicted genes. During a MAVID alignment of ancestral sequences in the progressive multiple alignment, position r from sequence 1 maps to position u in the ancestral sequence A, and position s maps to position v in the ancestral sequence B (solid lines). Even though sequence 3 is not in the multiple alignment yet, the constraint forces position u to be aligned before position v in the final multiple alignment (broken line). The constraint is enforced by removing all the matches violating the constraint from consideration during the anchoring of the alignment.











