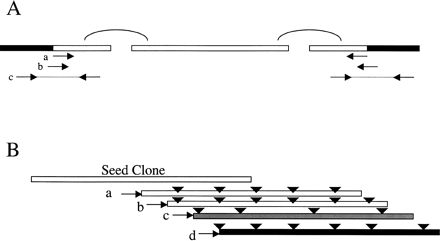
(A) Anchoring of the scaffold to the cloning end for each BAC clone. Each box represents a sequence contig obtained from the BAC assembly. Solid parts of the box indicate the cloning vector. Contigs were first linked into scaffolds using paired end reads, shown as curve connections in the figure. Three types of reads/clones were used to identify the clone junctions: (a) BAC end reads, (b) insert-vector junction reads, and (c) insert-vector junction clones. (B) Restriction enzyme digestion patterns of the candidate clones were used to filter out false positives caused by mismapping of the BAC end reads. The triangles represent restriction enzyme recognition sites. Clones a, b, and d share common sites, whereas clone c has a very different pattern. As a result, clone c is excluded from further analysis. Clone d is selected as having the minimal overlap.











