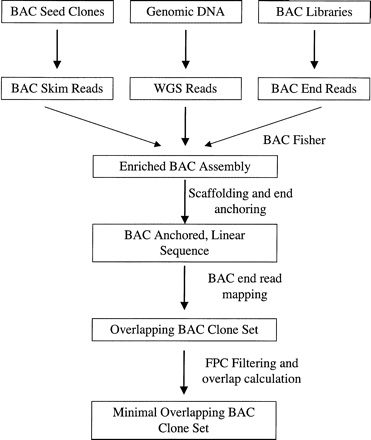
The CLONEPICKER pipeline. BAC skim reads from seed clones, WGS reads, and BAC end reads were used to establish the enriched BAC seed clone assembly using the ATLAS tools. Candidate BAC clones that overlap with the seed clone were first identified based on BAC end reads that mapped to the enriched BAC assembly. These candidate clones were then assessed by comparing their restriction enzyme digestion patterns. Clones that passed this filtering step were then analyzed for their overlap size with the seed clone. Clones with minimal overlap with the seed clones were selected as new clones for sequencing.











