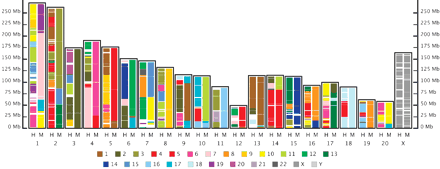
Rat/Mouse/Human comparison. Pash was used for pairwise comparison of the genome of rat with those of mouse and human. Chromosomes are shown with p-arm down. Each chromosome of the rat genome is represented as a pane (identified along the x-axis) containing two columns that have been “painted” using the Virtual Genome Painting method (http://www.genboree.org) according to similarity to chromosomes of mouse and human. Position within the rat chromosomes is indicated along the y-axis in megabases (Mb). Columns are colored according to similarity to human chromosomes (left columns, “H”), and mouse chromosomes (right columns, “M”). The same color code is used for similarity to both human and mouse, as indicated by the legend. For example, the first 25 Mb of rat chromosome 1 is similar to sequences from human chromosome 6 (indicated by pink coloring in the left column) and mouse chromosome 10 (indicated by yellow coloring in the right column). An interactive version of this figure is available at the Genboree site (http://www.genboree.org).











