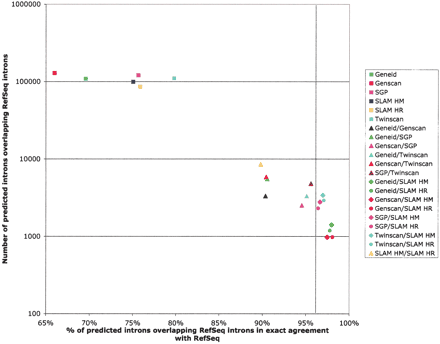
Enrichment for intron accuracy by gene prediction consensus. Intron accuracy of gene predictions by SLAM, other ab initio programs, and from consensus sets was measured by agreement with human RefSeq annotations. Comparative gene predictions utilizing two genomes were more accurate than those from noncomparative gene finders (Genscan and Geneid). Genscan, a noncomparative gene finder, had the greatest sensitivity, with close to 130,000 intron predictions overlapping RefSeq introns. Combining gene predictions to form consensus sets greatly increases accuracy while significantly reducing sensitivity. The vertical dotted line indicates the limit of the accuracy of consensus sets not involving SLAM.











