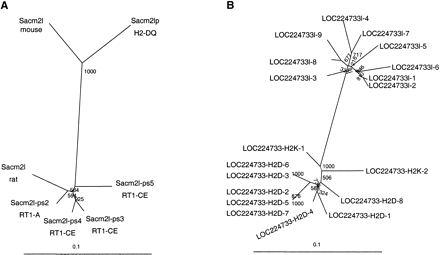
Independent evolution of proximal and first distal class I gene cluster in rat and mouse. (A) We detected a total of seven partial homologies to the Sacm2l gene in class I gene regions RT1-A and -CE, four of which match the 3′-end of Sacm2l (position 138894–139294). A similar transposition occurred in mouse leading to the presence of a Sacm2l-gene fragment in the H2-D/Q region (position 1554403–1554806 in NM_002588). Our phylogenetic analysis indicates that duplication and transposition took place after the speciation of mouse and rat. (B) Phylogenetic relationship between pseudogenes with homology to mouse gene LOC224733 in the MHC of rat and mouse. LOC224733-like sequences 1 and 2 are located in the rat A region; LOC224733-like sequences 3 to 9 map to rat CE (Fig. 2). The localization of 10 LOC224733-derived copies in mouse H2-K and H2-D is indicated. Species-specific clustering supports independent transposition and amplification of LOC224733 elements in rat and mouse. We calculated neighbor-joining trees as implemented in CLUSTALX. Bootstrap values (1000 replicates) are indicated.











