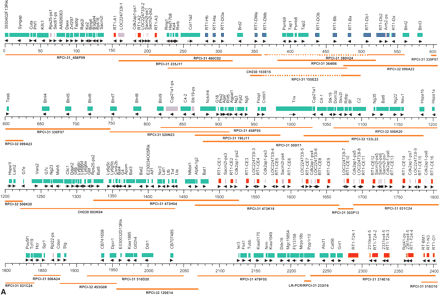
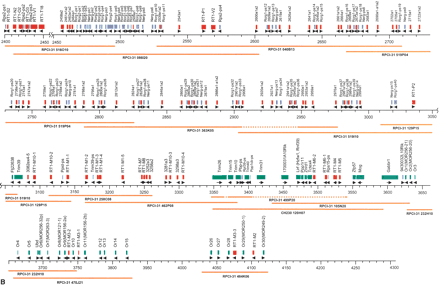
Physical map of the RT1 region including the positions of genes, pseudogenes, and gene fragments as detected by our sequence analysis. Nomenclature of non-class I and non-class II genes generally follows LocusLink (http://ncbi.nlm.nih.gov/LocusLink). Where no gene designation in mouse or human exists to date, accession numbers, Riken cDNA clone numbers, or EST designations are used. For brevity, Riken clone 5830469G19-related pseudogenes are designated as Rcrg1-ps, Riken clone 2210412D01-related pseudogenes as Rcrg2-ps, and EST 221625 (AI177969)-related pseudogenes as Nerg-ps. Class I gene fragments of the RT1-N and RT1-M1 regions are designated according to their position (kb) and the domains (a) or exons (ex) present. No unifying nomenclature exists for olfactory receptor genes. Therefore, the designation Or with consecutive numbers was used, leaving a gap for those Or genes that map between Or15 and Or26 according to the Baylor rat genome sequence. The mouse gene (MOR designation) was included in brackets where orthology was evident (see Fig. 5B). Red, class I genes and class I-derived gene fragments; blue, class II genes; purple, non-class I pseudogenes and non-class I gene fragments; green, framework genes (functional genes with exception of class I and class II genes). Map positions are indicated in kilobasepairs. For sake of clarity, the segment between 2450 and 3050 kb is shown at higher resolution (breaks indicated by pair of wavy lines). Sequenced clones are indicated with their library origin and plate coordinates. We used the RPCI-31 and RPCI-32 library resources (Woon et al. 1998). A minimal tiling path consisting of 38 BAC and PAC clones was derived from our previously constructed RT1 contig maps (Walter and Günther 2000; Ioannidu et al. 2001; Walter et al. 2002). Deletions within clones are displayed as dotted lines. We encountered RT1 intervals that were prone to deletions in a number of our clones, that is, between RT1-DMa and RT1-Db1 in the class II region, in interval G6f-Bat5 and proximal to Mog. For these regions, clones sequenced at low redundancy in the rat genome project at Baylor College were identified using BLAST and sequenced to high coverage for gap closure (BAC clones CH230–103E15, CH230–003K4, and CH230–120H07). The three library resources used each represent the rat RT1n haplotype. Clone RPCI31–484K06 containing class I genes RT1-M3-3 and RT1-M2 is connected to the major contig of 3.8 Mb by draft-quality sequence generated at Baylor College.











