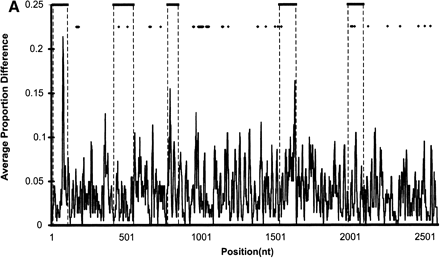
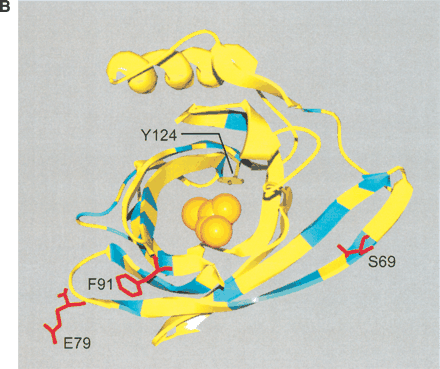
(A) Sequence divergence of rat α2u-globulin genes and pseudogenes. Genomic DNA from 22 rat α2u-globulin genes, including 12 suspected of being pseudogenes, identified as described in Methods. Nucleotide sequences were highly conserved (mean 93%±2% identity calculated from ungapped columns), and were aligned using HMMer (Eddy 1998) with manual adjustments. The proportion difference was calculated as the fraction of nucleotides in each column that differ from the consensus nucleotide. The average proportion difference from a sliding window of 10 nt is plotted against initial nucleotide position. Alignment columns with >50% gaps are shown as diamonds and were ignored for these calculations. Exons are shown as solid horizontal lines, with intron-exon boundaries defined by dashed vertical lines. (B). Site-specific KA/KS analysis of the MUP/α2u-globulin family, lipocalin homologs. ω+ codons with a posterior probability >0.90 by one codeml model, and >0.5 by at least one other model, are shown mapped to the crystal structure of murine MUP complexed with 2-(Sec-butyl) thiazoline (PDB 1MUP; Bocskei et al. 1992). Residues corresponding to ω+ codons are highlighted in blue; 2-(Sec-butyl) thiazoline is shown in orange. A single interior ω+ site corresponds to 1MUP:Y124 (shown in green). Although there are no data available to support a direct role in binding, this tyrosine residue neighbors, and has a similar orientation to, 1MUP:R126, which is required for thiazoline-binding (Bocskei et al. 1992). Y124 is as close to 2-(Sec-butyl) thiazoline as 1MUP:F60, a proposed ligand-binding residue (Darwish Marie et al. 2001). All mouse, and most rat, sequences contain a tyrosine in the position equivalent to 1MUP:Y124. However, two rat genes (mup_rn11 and mup_rn20) possess substitutions (to leucine and tryptophan, respectively) that might modify the geometry of the internal ligand-binding cavity. Consequently, these two genes might have evolved either to bind a ligand different from those of their paralogs, or else the same ligand with a different affinity. We also identified three codons (1MUP:S69, E79, and F91) that have been subject to rat lineage-specific positive selection. These are shown here in ball-and-stick format and colored red. These sites are the only alignment positions where amino acid substitutions are not conserved in rat α2u-globulins but are conserved in mouse MUPs. One rat α2u-globulin (mup_rn5) possesses unusual substitutions at all three positions (for S69, two substitutions at second and third codon positions [R→T]; for E79, a first position transversion [P→A]; and for F91, a second position transition G→E). mup_rn18 shares the R→T substitution at S69 with mup_rn18. This suggests that at least three sites in α2u-globulins may be under positive selection in the rat lineage alone, possibly reflecting the functional divergence of two genes (mup_rn5 and mup_rn18) from the other rat paralogs.











