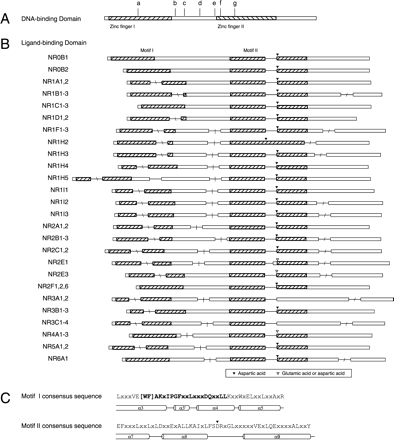
The gene structure encoding the DBD and LBD domains of the NR genes. Open bars are exons, drawn to scale; line segments, drawn at fixed length, give intron locations. (A) DBD splice junctions. Sequences are 75–78 aa in length. The shaded boxes indicate the location of the two C4 zinc finger motifs within this highly conserved domain. Introns may be found at seven different locations in the DBD across the entire family, or may be absent. Vertical hash marks indicate the location of junctions that were shared in the following groupings: a, NR2B, NR2C1,2; b, NR1A,1B,1C,1D,1F,1H4-5, NR5A; c, NR1I, NR4A; d, NR3A,3B,3C; e, NR2E3; f, NR2A, NR2F6; g, NR2E1; and not shown are group h, NR1H, NR2F, and NR6A, which have no intron in the DBD. (B) LBD splice junctions. Sequences are 170 (NR1D2) to 208 (NR0B1) aa in length. Each row is a schematic drawing giving the relative location of the splice junction and the group of NRs sharing the splice junction pattern. The position of splice junctions in orthologs was always the same, and thus species designations are omitted. Two conserved motifs (I and II, see text) in the LBDs are shown as the hatched areas. The location of a highly conserved negatively charged amino acid residue (aspartic acid or glutamic acid) in motif II is marked by an inverted triangle. The four regions within which introns were found are indicated by slash marks: “\” in motif 1, “|” intermotif region, no slash in motif II, and “/” after motif II (see text). (C) The consensus sequences of motifs I and II. The secondary structure of the corresponding part of the LBD, derived from crystallographic studies, is indicated below the sequence. Letters in bold correspond to the residues of the NR signature, involved in stabilizing the canonical fold of the NR LBDs (see Wurtz et al. 1996).











