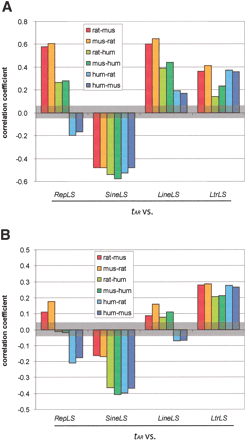
Pairwise correlations among measures of sequence divergence in comparisons among mammals. (A) Analysis of the original data. The amount of change by three processes was quantified in about 2500 1-Mb nonoverlapping windows genome-wide in two-way alignments among rat, mouse, and human. The function tAR is the substitutions per site in aligning ancestral repeats within each window, and RepLS is the portion of each window of the first (reference) species occupied by lineage-specific repeats. The portions occupied by lineage-specific LINEs, SINEs, and LTRs are given by LineLS, SineLS, and LtrLS, respectively. Correlation coefficients for each comparison are plotted. The brackets between roughly -0.05 and +0.05 denote the region in which the correlations lose statistical significance (P value more than 0.05) (B) Pairwise correlations among measures of sequence divergence in comparisons among mammals, after removing the effects of (1) G+C content in the first species, and (2) change in G+C content between the two species, using quadratic regressions.











