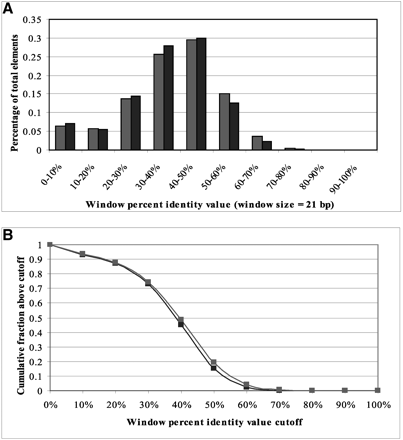
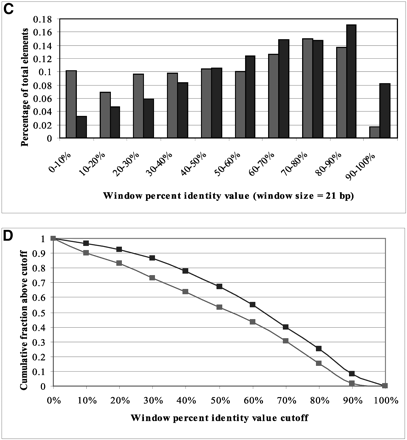
Conservation of known S. cerevisiae regulatory elements in yeasts. (A) Histogram of WPID values (21-bp window) of known S. cerevisiae regulatory elements (dark) and background sequences (light) within 1000 bp upstream of the translation start site between S. cerevisiae and S. pombe. (B) Cumulative distribution of WPID values. (C) Histogram of WPID values (21-bp window) of known S. cerevisiae regulatory elements and background sequences among S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus. (D) Cumulative distribution of the WPID values among the four yeast species. The conservation distributions of known elements and background are very similar between S. cerevisiae and S. pombe, without the good separation observed in the human–mouse comparisons. The distributions of known elements and background among the four yeasts are more differentiable than those of S. cerevisiae and S. pombe, indicating that sequences from multiple species can help identify regulatory elements.











