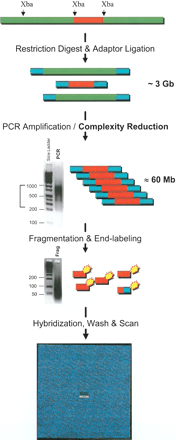
Complexity reduction assay and array hybridization. Sample genomic DNAs are digested with Xba I, and adaptors are ligated to the ends of restriction fragments. The fragments are then amplified by using one of the strands of the adaptor as a primer. Restriction fragments in the size range 250–1000 bp are preferentially amplified as shown in the gel image of PCR products. The narrow size range of amplicons is estimated to represent ∼60 Mb of sequence complexity, which is a 50-fold reduction in genome complexity. To allow efficient hybridization to 25-mer oligonucleotide probes on the array, the PCR products are fragmented with DNAse I. The size range of the fragmented PCR products is shown in the second gel image. The fragmented products are biotinylated and then hybridized to the arrays. Following a series of stringent washing and signal generation steps, the arrays are scanned; genotypes are then determined based on hybridization signal intensities.











