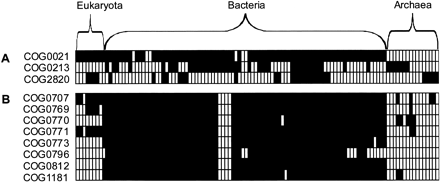
Phyletic patterns of the components of two functional modules. Each row is an orthologous group of genes, and each column is a species. Filled squares indicate that the orthologous group is present; blank squares indicate absence. Panel (A) shows the (deoxy) ribose phosphate metabolism pathway from EcoCyc. It has three components that evolve very flexibly: The observed deviation from the average number of module components per species is 0.682, whereas the random deviation is 0.677 for random shuffling and 0.802 for random sampling. The deviation when the module would behave evolutionarily perfectly modular would be 1.50. The modularity score of this pathway is thus 0.006 (random shuffling) or –0.172 (random sampling; see Methods). As the SD's of both types of random are respectively 0.033 and 0.074, this pathway falls within one SD of random and is thus as flexible as a random evolving group of genes would be. (B) The peptidoglycan biosynthesis pathway from EcoCyc. The modularity score of this pathway is 0.82 for random shuffling and 0.71 for random sampling; both are significantly more than random (more than 2 SD).











