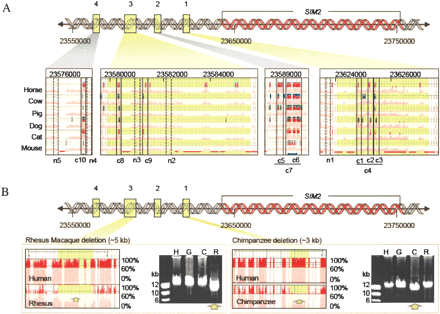
Noncoding sequences upstream of SIM2 chosen for functional characterization. (A) Conserved sequences (c1–c10) and nonconserved sequences (n1–n5) are located within four different intervals (coordinates based on NCBI contig NT_002836) (n6 is located ∼94 kb upstream). Visualization plots (see Fig. 1) show the species in which elements c1–c10 are conserved. (B) Conserved sequences deleted in the chimpanzee and rhesus macaque genomes. Comparison of syntenic human (H), gorilla (G), chimpanzee (C), and rhesus macaque (R) long-range PCR (LR–PCR) products by gel electrophoresis shows the deletion of interval 1 conserved sequences in chimpanzee genomic DNA and the deletion of interval 3 conserved sequences in rhesus macaque genomic DNA (yellow arrows). The visualization plots, generated by hybridization of the primate LR–PCR products to the human 21q arrays, indicate the positions of the human sequences (highlighted in yellow in A and B) deleted in the chimpanzee and rhesus macaque genomes. The deletions correspond to the drop in percent conformance, plotted on the vertical scale relative to the position in the human reference sequence. Green horizontal lines at the top of the plots indicate the positions of the LR–PCR products.











