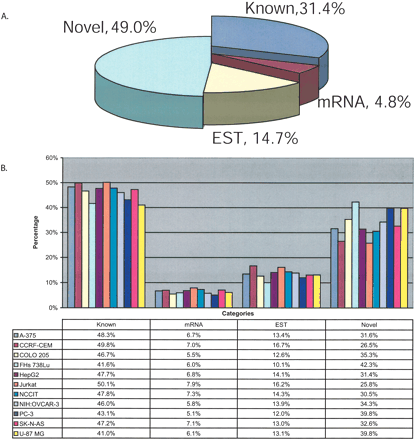
Characterization of all transfrags based on annotations. Transfrags from all 11 cell lines were classified based on their base pair overlap with annotations. The annotations are (1) Known, overlapping with known annotations (compiled by UCSC Genome Browser based on protein-coding genes from SWISS-PROT, TrEMBL, and TrEMBL-NEW); (2) mRNA, overlapping with mRNAs and not known; (3) ESTs, overlapping with ESTs and not known or mRNAs; and (4) Novel, not overlapping known exons, mRNAs, or ESTs. (A) Pie chart representing all transfrags from “1 of 11” map. (B) Bar graph shows the percentage of all transfrags from each of the 11 cell lines by annotation class. An overlap of even a single base pair is considered as an intersect between the transfrag and annotation.











