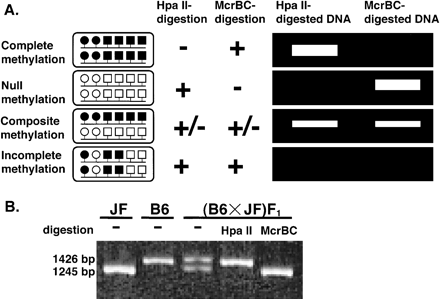
HpaII-McrBC PCR. (A) Principle of HpaII-McrBC PCR that distinguishes four different patterns in allelic methylation. (Left) The open and closed circles indicate unmethylated CCGG and methylated CmCGG sites, respectively. Similarly, the open and closed squares indicate unmethylated RC and methylated RmC sites, respectively. Each line with two circlesand four squaresindicateseach allele of genomic DNA. (Middle) The +, +/-, and - mean complete, incomplete, and no digestion with HpaII or McrBC, respectively. (Right) A schematic gel pattern of HpaII-McrBC PCR products in individual cases. (B) Proof of principle for HpaII-McrBC PCR. HpaII-McrBC PCR wasapplied to the intronic DMR of mouse Impact, a paternally expressed gene. JF, B6, and (B6 × JF) F1 indicate Mus musculus molossinus JF1, Mus musculus domesticus C57BL/6, and F1 hybrid generated between JF and B6, respectively. The PCR products from mock-treated (-), HpaII-digested, or McrBC-digested DNAs from JF, B6, or (B6 × JF) F1 were electrophoresed, stained with ethidium bromide, and visualized by UV illumination.











