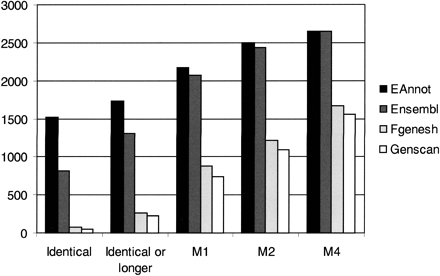
Comparison of transcripts predicted by EAnnot, Ensembl, Fgenesh, and Genscan with manual annotation. (Identical) Predictions with identical splice sites (coordinates, order, and number) compared with manual annotation; (Identical or longer) predictions with or including identical splice sites (coordinates, order, and number) compared with manual annotation; (M1) predictions missing a maximum of 1 splice site; (M2) prediction missing a maximum of 2 splice sites; (M4) predictions missing a maximum of 4 splice sites. Missing splice site can be due to incorrect splice site present or a predicted form being shorter than manually annotated form.











